Application of digital PCR and high-resolution melting method to find trace mutants in tumor samples
Summary:
Vogelstein and Kinzler (Proc., National Academy of Sciences, 1999) describe for the first time the concept of Digital PCR (dPCR), a method for identifying mutations in a small population of cells, such as primary tumor tissue. By diluting the sample DNA to a single molecular level, it can convert the PCR natural analog index into a digital signal. When PCR successfully amplifies these individual molecules, the product can be sequenced by observing the observed expected somatic homozygous mutation peaks, rather than a small number of low peaks in the background sequencing peak that are masked in the background peak. By amplifying amplifying products of a sufficient number of individual molecules, trace mutants can be identified based on the ratio of observed mutants to all successful amplification products.
method:
We used an improved dPCR method for the addition of HRM to find low levels of mutations in primary tumor samples using a special HRM profile.
Digital PCR requires a single DNA molecule as an amplification template, thereby making end detection a digital readout rather than a simulated DNA mixture. In order to apply HRM, more amplification products were needed to obtain a more reliable HRM profile set, so we chose five different dilution concentrations.
These diluted samples were based on subsequent sequencing methods with sensitivity close to 20% to detect trace mutants. HRM showed high sensitivity, and down to 2-5% of mutants were detected. Therefore, replication of a single mutant gene is sufficient to find their differences in the dissolution profile.
Fourteen different cancer genes were tested and 17 samples showed the presence of a possible minor mutant (12) or known mutation (5). (See Table 1)

For samples with known mutations, a mixture containing 5%, 2.5%, and 1.25% trace alleles was established. All samples were diluted at a target concentration of 5, 10, 20 and 40 copies, and the number of amplification cycles was 24.
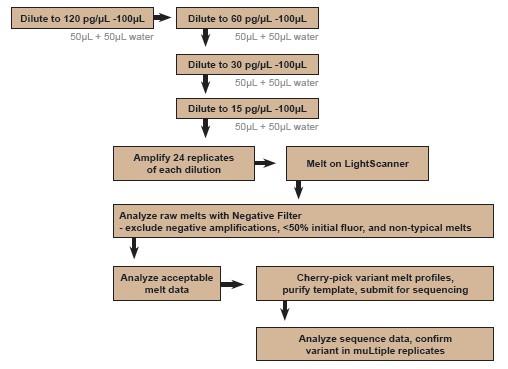
Samples without ideal amplification products were excluded from the high resolution melting curve analysis. Dilute samples showing possible mutations for sequencing. Figure 1 illustrates the workflow for applying HRM in this dPCR method.
result:
Nine of the 12 tumor samples were identified as having minor mutants. All five samples of known mutations were successfully identified at a dilution of 1.25%. Figure 2 illustrates that the "effective" concentration can be estimated by applying the probability of Table 2.

There are 6 amplification failures in this dilution of 5 copies, and it is recommended that the effective dilution concentration be close to 1-1.5 times (3-4.5 pg).

Figures 3 and 4 are an example of how to apply multiple dilutions on the same sample to determine the ideal dilution to identify trace mutants.


Figures 5a and 5b show that the known BRAFx15 mutations were diluted at 20 and 40 copies, respectively. The allele score reflects that the sequencing method correlates well with the dilution.

in conclusion:
These results show how the HRM pre-test improved dPCR method can be used to significantly reduce the work of subsequent sequencing stages to detect trace mutants compared to conventional dPCR.
In this study, identifying candidate HRM curves reduced the workload by 90% compared to traditional template resequencing methods.
Ecg Holter,Holter Ecg Machine,Bluetooth Ecg Machine,Portable Holter Ecg Machine
Changchun Yingwang Times Digital Co., Ltd. , https://www.ccsdsm.com